Probleemstelling:
Membrane targeting may play a pivotal role in human disease. Tau protein is an intrinsically disordered protein that interacts with the cell membrane, inside the neuron and it is involved in neurodegenerative disease [1]. As Tau protein interacts with anionic lipids, its structure is altered and it becomes a compacted, partially folded protein, which may be the catalyst for Tau aggregation into disease causing neuroï¬brillary tangles. Additionally, it interacts hydrophobically with the core of the membrane, causing disorganization of the lipids and destabilizing the membrane structure, which may also be associated with a disease mechanism [2]. A tendency of this protein for the formation of ion channels allows the passage of nonspeciï¬c ions [3]. This reinforces the role of Tau in the balance of K+ ions within the neuron and in the electrical signal propagation between cells. K+ and Na+ imbalances in neurons were observed in Alzheimer’s disease brains [4]. This fact could arise from the malfunctioning of several proteins, including Tau, which causes changes at the structural and signaling level, affecting the normal concentrations of these ions. However, little information is provided concerning the effect of this protein on membrane and its function. In this research work, the knowledge of this protein’s structure and function will be extended. To achieve this goal, the interaction between Tau protein and the membrane will be simulated at the atomic scale, with and without a gene mutation in the Tau protein. This will shed a light on how the interaction with the membrane is affected by the gene mutation.
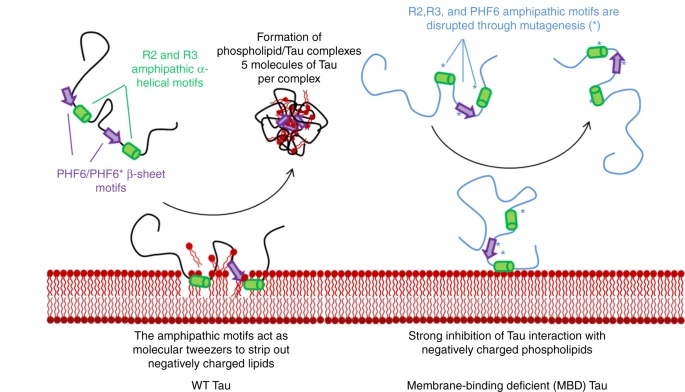
Fig 1. Schematic picture showing Tau and negatively charged phospholipids interaction. The interaction between Tau monomers and negatively charged phospholipids in vesicles is seen after Tau changes to fibrils. This interaction potentially takes place when short α-helical (green) and β-sheet (purple) motifs are formed. This might be followed by the mechanism (left) of lipid molecules being segregated into protein/phospholipid complexes which are highly stable. Regarding these results, mutants were designed, being capable of forming fibrils yet not being able to interact with phospholipids (right) to make protein/phospholipid complexes. This design will be applied in this thesis to facilitate the investigation of membrane binding effects on the normal and pathological functions of Tau. Figure is adapted from reference [5].
Experimental techniques have come a long way to probe structural and dynamical information at multiple scales. However, Due to the fluid nature of the membrane and the reversibility of protein–membrane interactions, the experimental study of these systems remains a challenging task. Computational biology provides a bridge to understand experimental results at the molecular level, makes predictions that have not been seen in vivo, and offers a suitable approach to study protein–lipid interactions. This thesis will use molecular dynamics (MD) simulations to study the membrane-protein interaction.
Speciï¬c lipid–protein interactions are essential components in signaling, cell division and cell structure. The majority of MD simulations of biological membranes and membrane proteins have been limited to homogeneous bilayers of a single lipid type. These bilayers, however, do not emulate realistic biological membranes, as other membrane components play essential roles in not only maintaining membrane integrity and properties, but also in its function. Consequently, the information these studies can provide on protein-lipid interactions is somewhat limited. The membrane environment (fluidity and order) and lipid composition are determinant for the interaction of this peripheral protein with a bilayer [6]. In this research, we will construct a complex lipid bilayer that mimics the composition of biological membranes consisting of many different types of lipid [7,8].