Probleemstelling:
Viruses may enter and infect the cells by membrane fusion. This process involves fusion peptides, which are short amino acid chains (peptides), that interact with the membrane to initiate the fusion. As an example, the fusion peptides of viral spike protein assist the corona virus to enter the host cell. This behavior has inspired a new approach in drug design. It is often a challenge to bring the less soluble drug molecules into the interior of the cells. A particularly interesting route of thought is then to attach the drug molecule as 'cargo' to a cell penetrating peptide (CPP), a short amino acid chain (peptide) that can permeate membranes. Potential clinical applications of this facility have prompted enormous effort, both experimental and theoretical, to better understand how CPPs manage to overcome the prodigious thermodynamic cost of membrane permeation. Current use is limited by a lack of cell specificity, meaning that drugs-loaded CPP cannot distinguish for instance between healthy and cancerous cells. There is currently an insufficient understanding of the modes of their uptake, which hinders developments to improve the specificity e.g. by varying the amino acid sequence, charges, and chain length in a guided manner. Modeling at the molecular scale can aid in understanding the process.
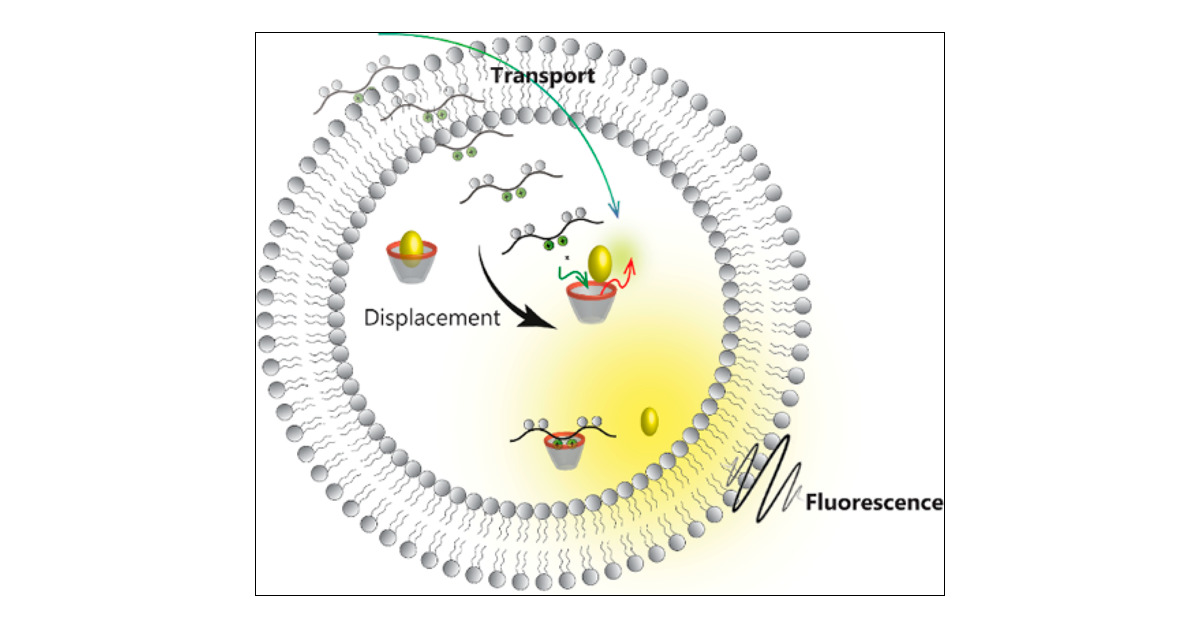
Figure 1. A cell penetrating peptide (CPP) is a short chain of amino acids that can enter the cell and that can carry a drug across the membrane. The route that will be investigated in this thesis is the direct transit through the membrane (translocation).
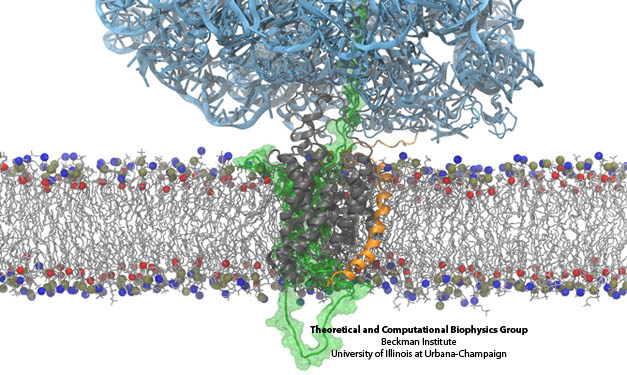
Figure 2. A protein (light blue) is attached to a membrane (horizontal in figure) consisting of phospho-groups (blue and red atoms) and hydrocarbon chains (light grey). Several peptide chains (dark grey and orange) are immersed in the membrane. Water atoms are not shown.